TL:DR previous part
I was approached by one of my clients with a work that required to parse almost 500 million European routes to see what routes can be traveled by land transport. After some research, i decided to use graphhopper server application and python celery library.
Data preparation
Graphhopper uses osm.pbf files as input files in order to correctly draw its maps and compute directions. These files can be downloaded from geofabrik site and can be freely used. However, another issue occurred during the preparation step: file sizes. The latest Europe map weighs more than 22Gb and that’s the compressed file! Graphhopper will also extract and set up his own metadata which will require additional storage usage. Even more, the larger the osm file the more there requirements for the RAM. That’s a huge requirement for memory, so i decided to shrink this osm file as much as possible. I needed only the West Europe map without Eastern Europe, Scandinavia, and England.
Shrink OSM files
Fortunately, osm files can be easily manipulated and feated to your needs. There is a tool called [osmosis] (https://wiki.openstreetmap.org/wiki/Osmosis) that can do that and much more. Here is an example of extracting a map of Germany with supplied polygon into a separate map file:
$: osmosis --read-xml file="planet-latest.osm" --bounding-polygon file="country2pts.txt" --write-xml file="germany.osm"Where country2pts.txt is a polygon filter file. In order to create such polygon one can use an online tool called hotosm. The polygon file itself is just a collection of map points on each row that describes our polygon boundaries:
australia_v
first_area
0.1446693E+03 -0.3826255E+02
0.1446627E+03 -0.3825661E+02
0.1446763E+03 -0.3824465E+02
0.1446813E+03 -0.3824343E+02
0.1446824E+03 -0.3824484E+02
0.1446826E+03 -0.3825356E+02
0.1446876E+03 -0.3825210E+02
0.1446919E+03 -0.3824719E+02
0.1447006E+03 -0.3824723E+02
0.1447042E+03 -0.3825078E+02
0.1446758E+03 -0.3826229E+02
0.1446693E+03 -0.3826255E+02
END
second_area
0.1422436E+03 -0.3839315E+02
0.1422496E+03 -0.3839070E+02
0.1422543E+03 -0.3839025E+02
0.1422574E+03 -0.3839155E+02
0.1422467E+03 -0.3840065E+02
0.1422433E+03 -0.3840048E+02
0.1422420E+03 -0.3839857E+02
0.1422436E+03 -0.3839315E+02
END
ENDUsing hotosm i created .geojson file and used its boundaries to create such polygon filter file. After that i only needed to extract land routes in the creared custom polygon:
$: osmosis \
--read-xml europe-latest.osm \
--way-key-value keyValueList="railway.tram,railway.tram_stop" \
--bounding-polygon file="created_polygon_filter_file_.txt"
--used-node \
--write-xml city_tram.osmBecause in this task i had only a list of cities to create routes permutations with i also needed to determine the exact coordinates for each city. As now i had our custom Europe polygon i used shapely in order to check how properly the coordinates were detected:
import shapely.geometry
import shapely.geometry.polygon
# Points from our custom polygon
coords = [
(81.434750, -5.863332),
(74.786230, -6.704456),
(62.807440, -34.492960),
....
(81.434750, -5.863332)
]
CUSTOM_EUROPE_POLYGON = shapely.geometry.polygon.Polygon(coords)
with open(path, 'r') as input_file:
for coordinates in csv.DictReader(input_file):
if not CUSTOM_EUROPE_POLYGON.contains(shapely.geometry.Point(coordinates['lat'], coordinates['lng'])):
print('Skipped: {}, not in europe poly'.format(term))
continueI now had an osm file which weighted in 12 Gb and that was an uncompressed osm file. The next step was the server setup.
Server setup
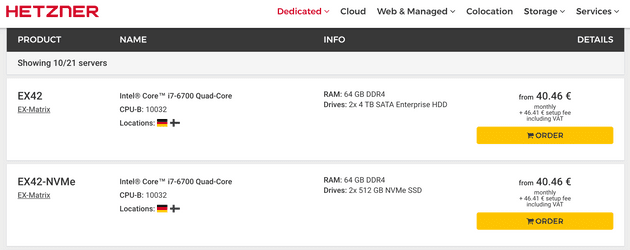
The result osm file weighted 12 Gb uncompressed, graphhopper will unwind it and add an additional 12 Gb for its metadata, i also needed a good amount of memory in order to comfortably work with routes. The list itself had almost 500 million routes to parse, so I could not parse it in one go, so i really needed to track what routes were processed and what not. In order to do that i decided to use redis server, its simple, reliable and very fast, also it can be used with python celery library which i decided to also use in my setup. After some considerations i decided to use hetzner EX42 type box.
Workers
I decided to use celery and redis as transport. The script will read each term and start to combine it in a loop with other terms until CONCURENCY number of routes is collected, then it will pass them to the celery worker group and wait for the processing to complete after that receive the results from celery and write them down. I also decided that it need a config option to start from a given point in the file, so i have introduced START_POINT config env variable.
import requests
import csv
import os
import itertools
import concurrent.futures
import router.redis_client
import celery
import hashlib
from router.tasks import lookup_directions
import router.hasher
START_POINT = int(os.environ.get('START_POINT') or 0)
CONCURENCY = 500
MAX_RESULT_BATCH = 10_000
def flush_batch_main(batch):
execution_result = celery.group(lookup_directions.s(
row[0], row[1]) for row in batch)().get()
return [[], execution_result]lookup_directions is our celery task and is pretty straightforward:
from __future__ import absolute_import, unicode_literals
from celery import Celery
# Use redis as broker
app = Celery('router',
broker='redis://',
backend='redis://',
include=['router.tasks'])
GRAPH_HOPPER_URL = 'http://localhost:8989/route?point={}&point={}&type=json&locale=en-US&vehicle=car&weighting=fastest&elevation=false&key='
# Reuse request socket and config between the requests, increases the performance
requests_session = requests.Session()
@app.task
def lookup_directions(start_point, end_point):
path = '{} {}'.format(start_point[0], end_point[0])
try:
result = requests_session.get(GRAPH_HOPPER_URL.format(
start_point[1], end_point[1])).json()
if result['paths'] and result['paths'][0]:
first_path = result['paths'][0]
return {
'merged_term': path,
'term_start': start_point[0],
'term_end': end_point[0],
'coordinates_start': start_point[1],
'coordinates_end': end_point[1],
'distance': first_path['distance'],
'time': first_path['time'],
'hops': len(first_path['instructions'])
}
except Exception as identifier:
print('Skipped, error: {}'.format(identifier))
return {
'merged_term': path,
'term_start': start_point[0],
'term_end': end_point[0],
'coordinates_start': start_point[1],
'coordinates_end': end_point[1],
'distance': None,
'time': None,
'hops': None
}The format of graphhopper api can be looked here. The main script looked like this:
def main(input_file, result_file, flush_batch, hasher, concurency, max_result_batch):
writer = csv.DictWriter(result_file, fieldnames=['merged_term', 'term_start', 'term_end', 'coordinates_start', 'coordinates_end', 'distance', 'time', 'hops'], quoting=csv.QUOTE_ALL)
writer.writeheader()
terms = list(csv.reader(input_file))
row_batch = []
result_batch = []
i = 0
for row_1 in terms[START_POINT:]:
for row_2 in terms:
# Track the current permutation number
if i % 1000 == 0:
print(i)
i += 1
sorted_points = [row_1,row_2]
sorted_points.sort(key=lambda x: x[0])
start_point = sorted_points[0]
end_point = sorted_points[1]
path = '{} {}'.format(start_point[0], end_point[0])
if start_point[0] == end_point[0] or route_exists(path):
print('Skipped, already exists: {}'.format(path))
continue
row_batch.append([start_point, end_point])
if len(row_batch) >= concurency:
row_batch, execution_result = flush_batch(row_batch)
result_batch.extend(execution_result)
if len(result_batch) >= max_result_batch:
print('Flushed')
writer.writerows(result_batch)
for entry in result_batch:
memorize_route(entry['merged_term'])
result_batch = []
if row_batch:
row_batch, execution_result = flush_batch(row_batch)
result_batch.extend(execution_result)
if result_batch:
writer.writerows(result_batch)
for entry in result_batch:
memorize_route(entry['merged_term'])
return True
if __name__ == '__main__':
with open(os.environ['INPUT'], 'r') as input_file:
with open(os.environ['RESULT'], 'a+') as result_file:
main(input_file, result_file, flush_batch_main,
router.hasher, CONCURENCY, MAX_RESULT_BATCH)memorize_route and route_exists is just a wrapper for redis client that checked redis key for existence and write one after we process it. Everything was in place and i was ready to start the processing. However, new challenges awaited me ahead. Stay tuned for the final article!